Scientific data stored in Matlab files can be easily imported into sigclear platform. As an example, we show how to use sgloadmat to import the UV and circular dichroism (CD) spectra from a matlab data file.
The matlab file of the CD-UV spectra can be download here. For the analysis of DNA conformation structure, the UV and circular dichroism (CD) spectra at different temperatures are measured in a laboratory. The temperatures, wavelength range, UV spectrum and CD spectrum are stored as the following variables in the matlab file:
- T temperature, of 24X1 8-bits integers
- wavelength wavelength, of 101X1 32-bits integers
- melt_cd_water CD spectra at the 24 temperatures, of 101X24 double floats
- melt_uv_water UV spectra at the 24 temperatures, of 101X24 double floats
load the spectra in command line
load a vector variable
A 1D vector variable can be loaded as scalar or vector field by sgloadmat for different purposes:- load a column vector as scalar field
- sgloadmat temperature=T < dna_data.mat > data.sg
- load a column vector as vector field
- sgloadmat temperature=T.T < dna_data.mat > data.sg
load a matrix variable
In matlab, matrix A of mXn dimension is stored as A[1][1], A[1][2],..., A[1][n], A[2][1],...,A[2][n],... By defaults, the A will be imported as a vector of n samples in m columns. For example,
- sgloadmat cd=melt_cd_water < dna_data.mat > data.sg
- sgloadmat cd=melt_cd_water.T < dna_data.mat > data.sg
load multiple variables
Multiple variables can be loaded simultaneously as
- sgloadmat cd=melt_cd_water.T uv=melt_uv_water.T temperature=T < dna_data.mat > data.sg
- cd vector field of 101 samples
- uv vector field of 101 samples
- temperature scalar field
or
- sgloadmat cd=melt_cd_water uv=melt_uv_water wavelength=wavelength < dna_data.mat > data.sg
- cd vector field of 24 samples
- uv vector field of 24 samples
- temperature scalar field
export to matlab file
sgoutmat automatically stores all fields to variables in a matlab file.
- sgoutmat < data.sg > data.mat
Please notice matlab needs memory to load a full matlab file. Due to the SOIG design, sigclear platform can handle datasets much larger than the memory, but the huge matlab data file may lead to memory problem for matlab.
load the spectra in SCons script
More conveniently, the CD-UV spectra can be load in SCons script asfrom experiment import *
Process('data', 'DATA/dna_data.mat',
'''
sgloadmat cd=melt_cd_water.T uv=melt_uv_water.T temperature=T
| sgattribute s0=230 index=temperature
''')
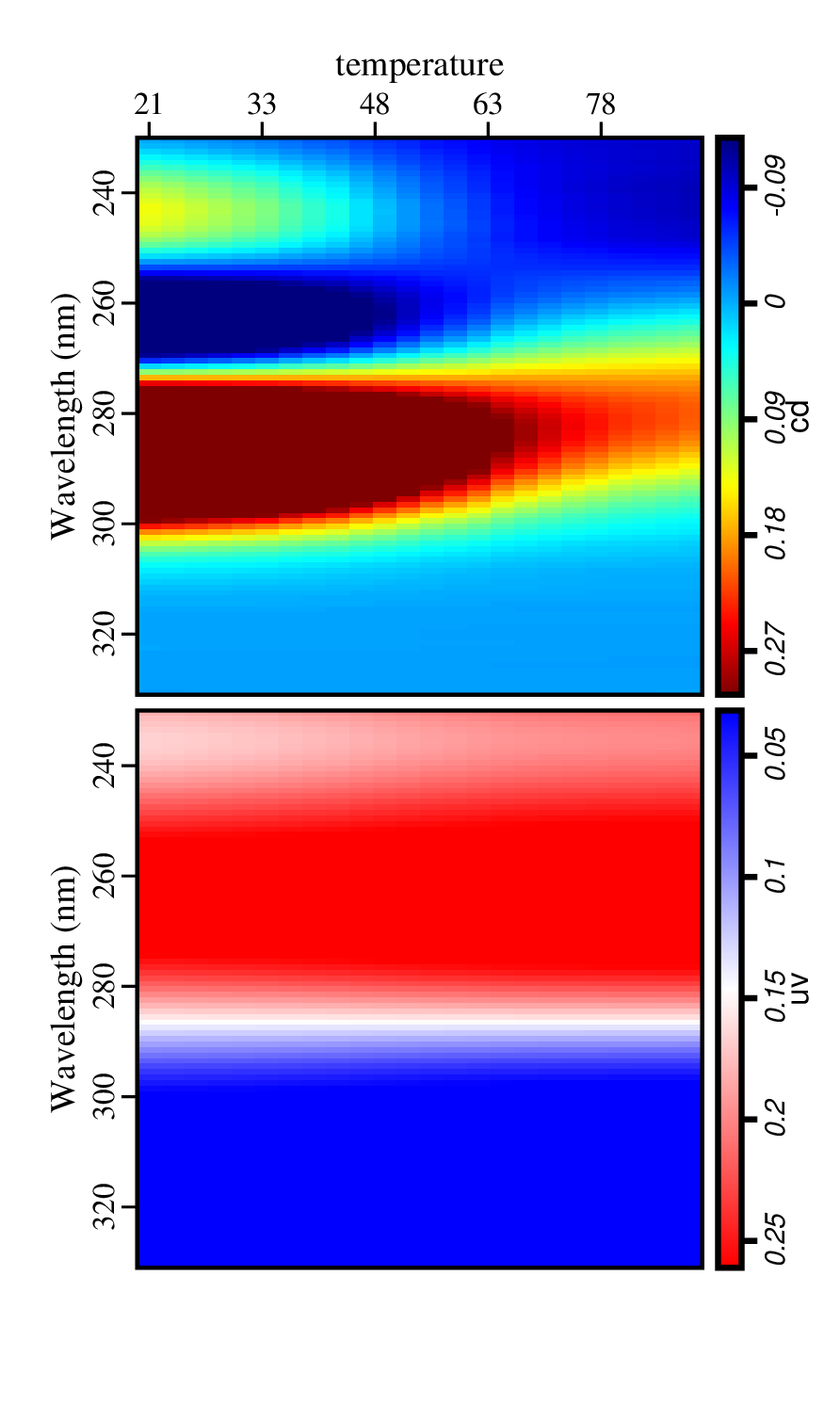
Figure('./data.png',
'''
sgplotps left.label="Wavelength (nm)"
uv.image=bwr cd.image=rainbow
''')
References:
- Jaumot, Joaquim; Escaja, Nuria; Gargallo, Raimundo; Gonzalez, Carlos; Pedroso, Enrique; Tauler, Roma. "Multivariate curve resolution: a powerful tool for the analysis of conformational transitions in nucleic acids." Nucleic Acids Research 30(17), e92, 2002.
- Download the matlab CD-UV spectra
